Nutrient utilization pathways in Mtb
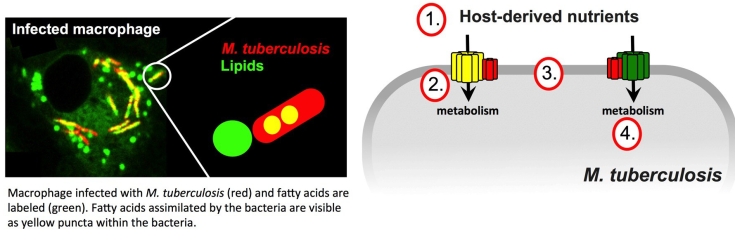
Pathogenic bacteria have evolved highly specialized systems to extract and utilize nutrients from their hosts. During infection Mtb preferentially scavenges lipid nutrients and the bacterium’s capacity to assimilate and metabolize host-derived lipids is considered a defining characteristic of this pathogen.
Mtb primarily metabolizes lipids (fatty acids and cholesterol) to produce energy and maintain persistent infection. A large proportion of genes in the Mtb genome are dedicated to lipid metabolism but the most of these genes and gene products remain uncharacterized. We are using genetic approaches to elucidate the critical proteins and pathways that are involved in the assimilation and metabolism of lipids and other nutrients by Mtb during infection. Additionally, we are using high throughput approaches to identify small molecules that perturb these pathways for drug development.
We are assigning function to uncharacterized proteins in these pathways and we are finding that multiple Mtb proteins are required for the utilization of different nutrients. We are actively perusing small molecule inhibitors that target these “bottlenecks” as a way to chemically block multiple metabolic pathways simultaneously.
 We are interested in the following questions:
We are interested in the following questions:
- What nutrients are scavenged from the host and how does the bacteria assimilate these nutrients?
- How are host-derived nutrients imported across the bacterial cell envelope and what bacterial proteins are required for this?
- How are the nutrient uptake systems regulated to coordinate the uptake of specific nutrients at different stages of disease, and if so, how?
- What is the fate of the nutrients once assimilated by the bacterium and how does the bacterium coordinate the simultaneous utilization of multiple nutrients?
Chemical-genetics of Mtb infection
Mtb replicates within host macrophages and this contributes to Mtb’s success as a pathogen. To survive within macrophages Mtb relies on a specialized metabolism, perturbs host cell functions, and uses detoxification pathways to counter the immune-mediated stresses associated with the intracellular environment (i.e. low pH, ROI and RNI).
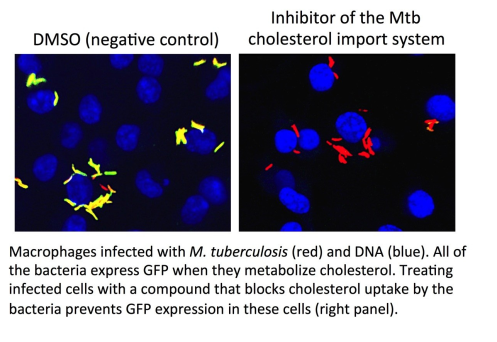
Recently we have identified numerous small molecule inhibitors that inhibit intracellular replication of Mtb. These compounds either perturb bacterial virulence/survival pathways or promote a more robust anti-Mtb innate immune response. We are interested in understanding the specific mechanism of action for these inhibitors against Mtb during infection in macrophages, and during in vivo infection in the mouse model of TB. These studies are revealing novel molecular targets that are “druggable” in the context of infection and are shedding new light on fundamental questions in the area host-pathogen interactions.
To identify targets of small molecules that inhibit Mtb replication in the context of the infection we are developing novel cell biological, immunological, and genetic methodologies. We are finding that bacterial targets of these compounds are not “classically” regarded as drug targets in Mtb. We are interested in evaluating drug combinations containing these compounds as a way to shorten TB chemotherapy.
We are interested in the following questions:
- What survival or virulence pathways in Mtb can be chemically perturbed to reduce bacterial survival during infection in macrophages?
- How can the immune response be chemically modulated to shorten or improve the outcome of current TB chemotherapy?
- Can drug discovery and target identification in the context of infection be improved and what is the therapeutic potential of these inhibitors?
Mtb physiology in necrotic granuloma tissues
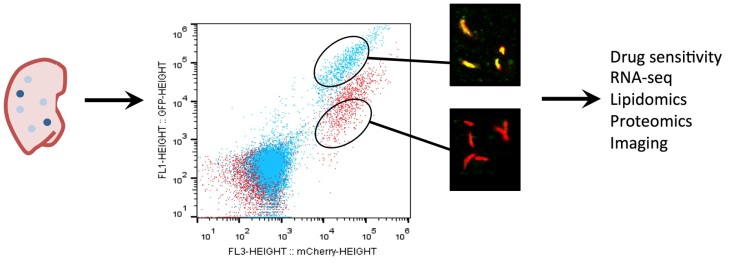
The physiology of Mtb within the tissue environment in a granuloma lesion is an enormous black box in our understanding of TB disease. Bacteria sequestered in granulomas are recalcitrant to antibiotic treatment and immune-mediated clearance. The 4-6 month long “continuation phase” of TB chemotherapy is needed to eliminate these bacteria and prevent disease relapse. Thus, understanding the physiology of Mtb in granulomas may help lead to the discovery of small molecules that target this recalcitrant population of bacteria. Additionally, it is thought that bacteria within these lesions develop a drug tolerant phenotype, which makes treatment less effective and promotes the evolution of drug resistance in Mtb.
We are interested in understanding the physiology of Mtb within different granulomas. To study Mtb within these environments we use laser confocal imaging and are developing sub-population methodology to quantify aspects of Mtb physiology in vivo. We are also applying genetic approaches to identify mutants that specifically do not survive within particular granuloma lesions. To facilitate these studies we are using alternative mouse models of TB infection that develop a spectrum of tractable granulomatous lesions. We are asking questions around how the physiology of Mtb in these lesions impacts drug efficacy and what bacterial pathways are most critical for survival in this tissue. We predict that these studies will shed light on Mtb in this important phase of disease and will reveal new targets for drug development.
We are interested in the following questions:
- What pathways in Mtb are specifically required for bacterial survival in different granulomas? Can we discover drugs that inhibit these pathways?
- Does the physiology of Mtb within granulomas contribute to drug tolerance, if so, how?
- How do particular immune cells or responses impact Mtb physiology in the context of different granulomas and tissues?